There is mention in the R-mark code base of "Parm-specific" settings in the mark.make.model function line 1234 (I didn't make up that line number), but this was really referring to parameter classes (e.g. p, phi), not link functions within a parameter class.
I would like to use both a log and logit link to model the survival parameter. We have migrating salmon which typically swim in a fair direct path between detection points. We would like to estimate a survival rate per unit time.
Let
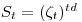 be a survival rate function, where
be a survival rate function, where 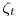 is the survival rate per week, and
is the survival rate per week, and 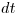 is the time in weeks to travel between detection points (aka occasions).
is the time in weeks to travel between detection points (aka occasions). Using a log link we get the following
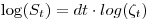
where
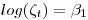 in a regular mark beta parameter in a model without an intercept allowing us to estimate survival rate as
in a regular mark beta parameter in a model without an intercept allowing us to estimate survival rate as 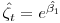 , given the invariance properties of MLE's.
, given the invariance properties of MLE's.Now the problem is we have subgroups, one of which pauses in one to two of the segments during migration, for this group*segment we would like to use a logit link function and just estimate the probability of survival, while in other segments we would like an estimate a survival rate.
For now I am just using a log link and a separate beta parameter with '1' in the design matrix for these special segments, giving us
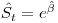 as our estimator, but as you know there is no bounds imposed when using the log link. Occasionally, we get some approx 95% CI that reveal the approximate nature as survival in this segment is quite high. Would a journal accept an upper bound of 1.05? Talk about great survival!!
as our estimator, but as you know there is no bounds imposed when using the log link. Occasionally, we get some approx 95% CI that reveal the approximate nature as survival in this segment is quite high. Would a journal accept an upper bound of 1.05? Talk about great survival!!Am I stuck using GUI MARK or have I missed something somewhere? Please feel free to berate as necessary.